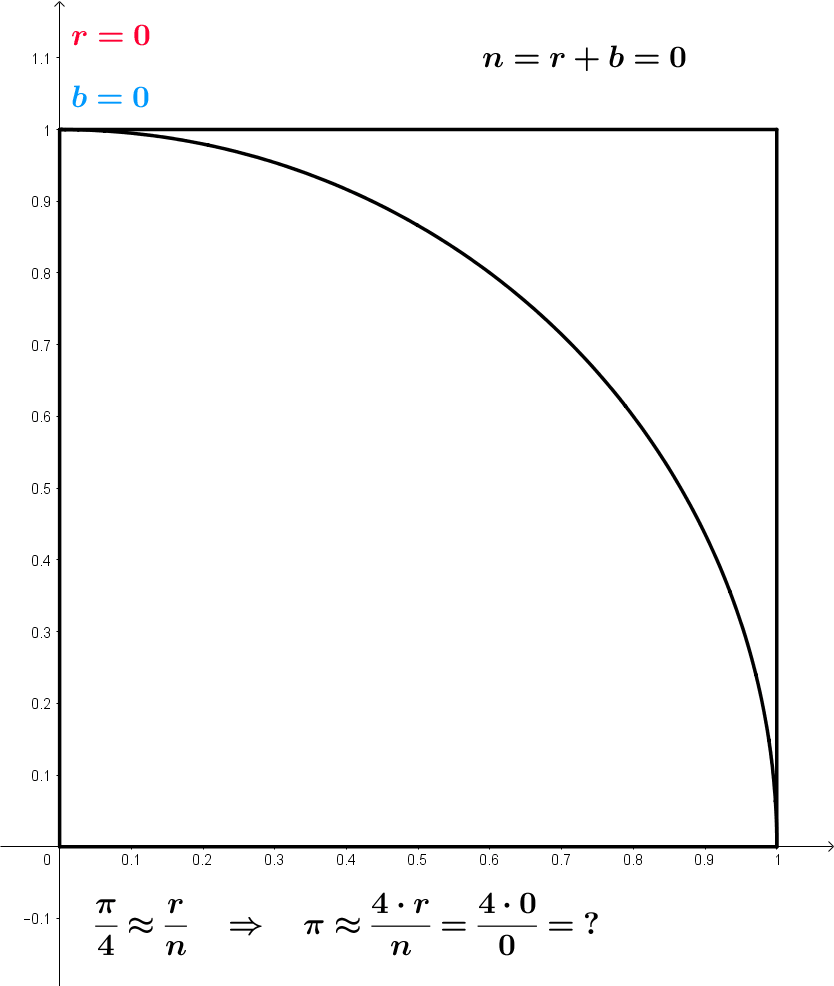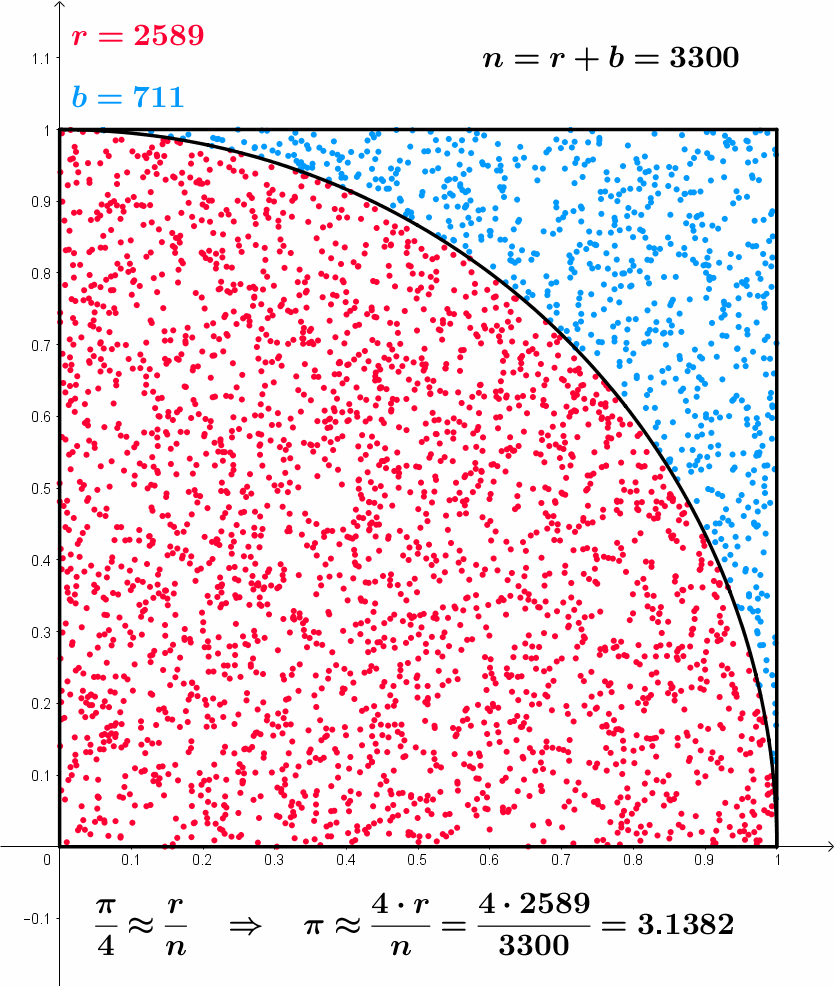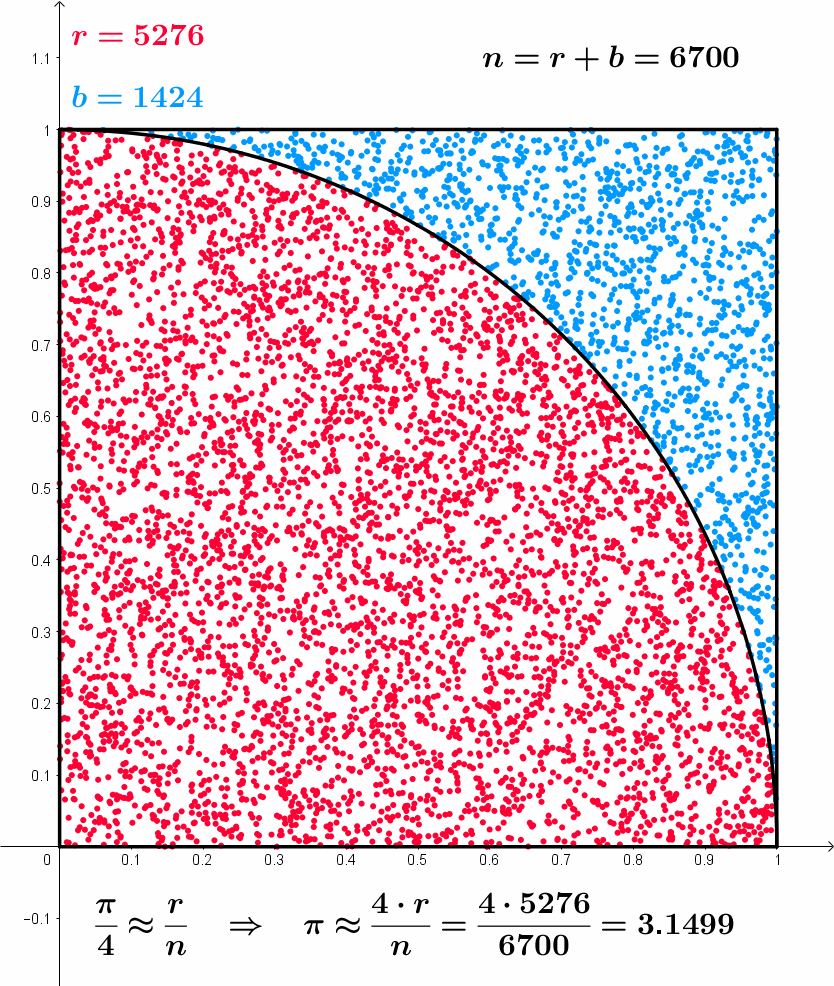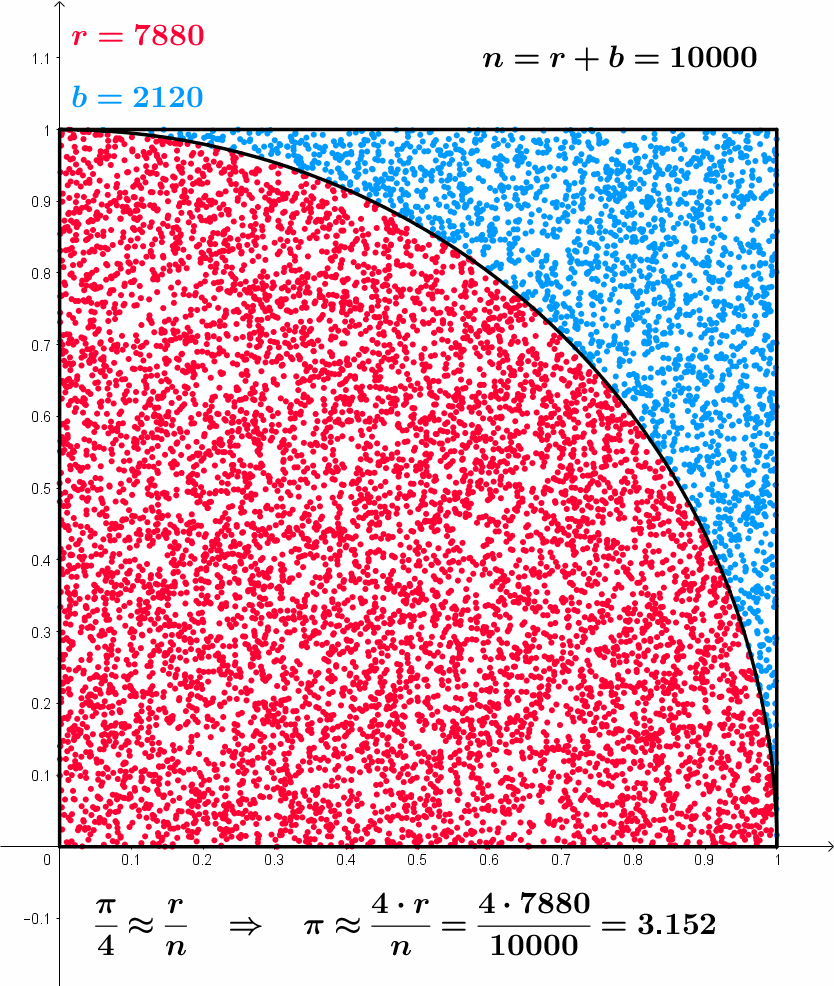
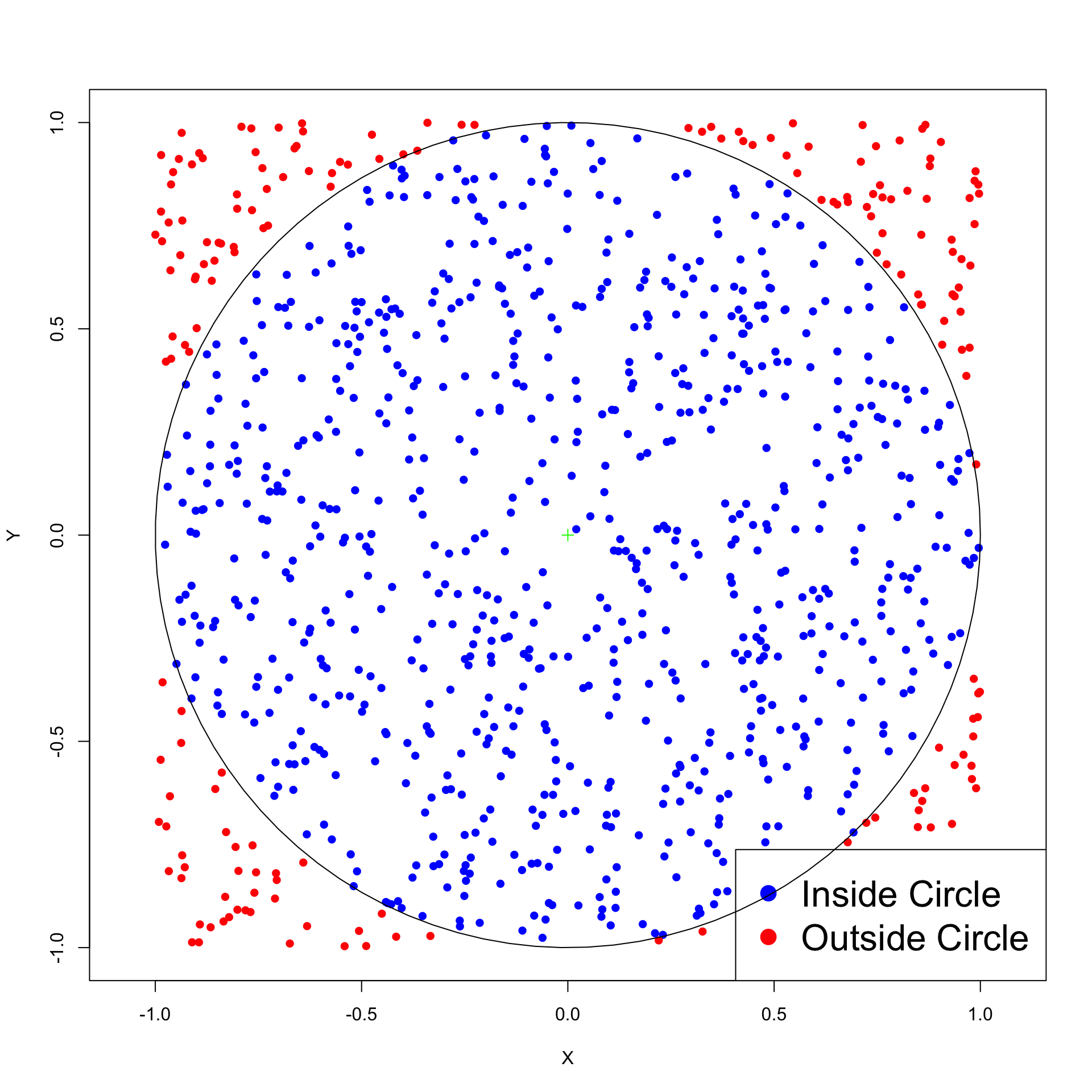
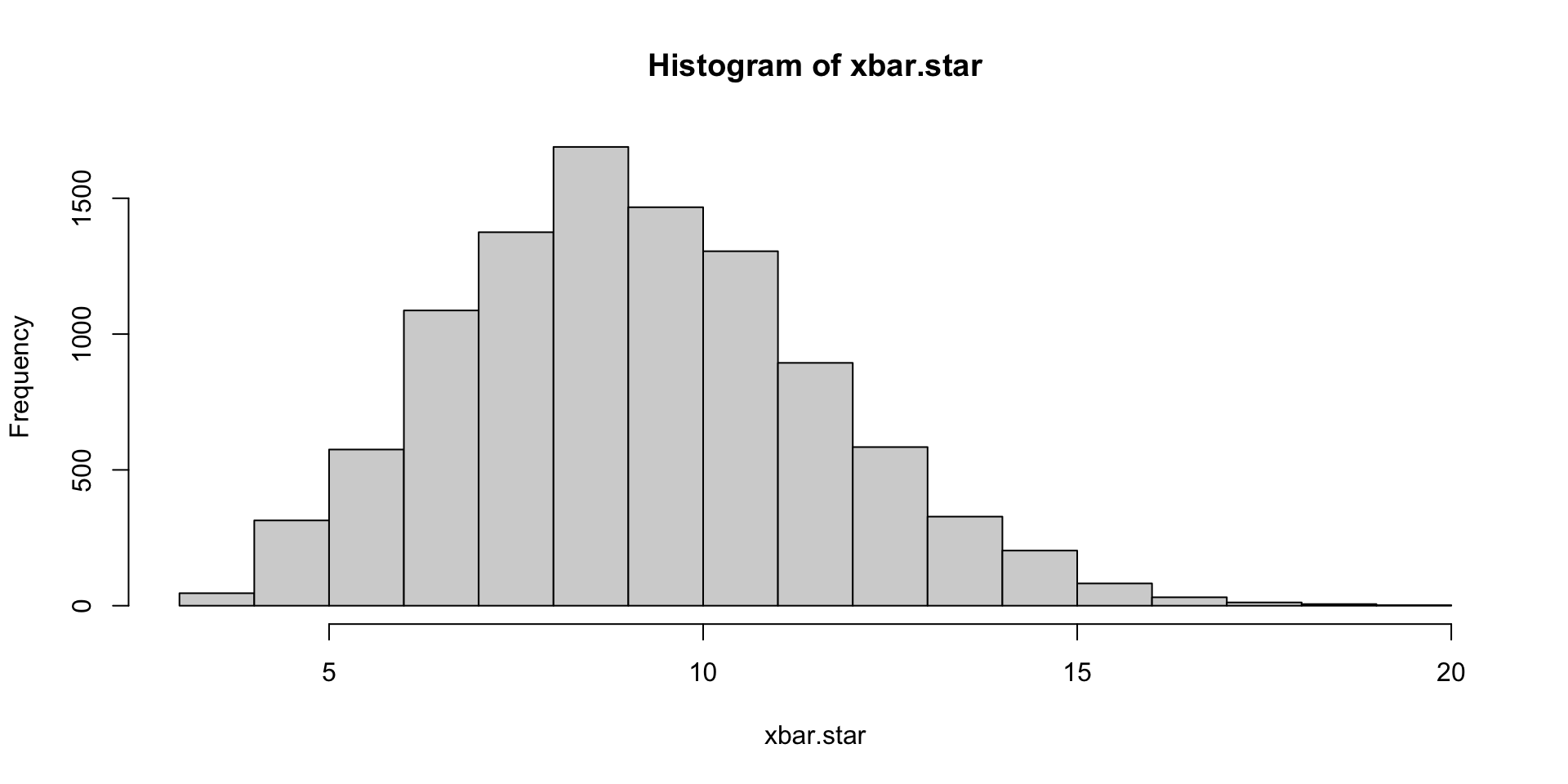
After 10000 iterations pi is 3.14560000, error is 0.00400735
After 20000 iterations pi is 3.13260000, error is 0.00899265
After 30000 iterations pi is 3.13253333, error is 0.00905932
After 40000 iterations pi is 3.14170000, error is 0.00010735
After 50000 iterations pi is 3.14248000, error is 0.00088735
After 60000 iterations pi is 3.14186667, error is 0.00027401
After 70000 iterations pi is 3.14200000, error is 0.00040735
After 80000 iterations pi is 3.14155000, error is 0.00004265
After 90000 iterations pi is 3.14177778, error is 0.00018512
After 100000 iterations pi is 3.14084000, error is 0.00075265Main Bootstrap idea
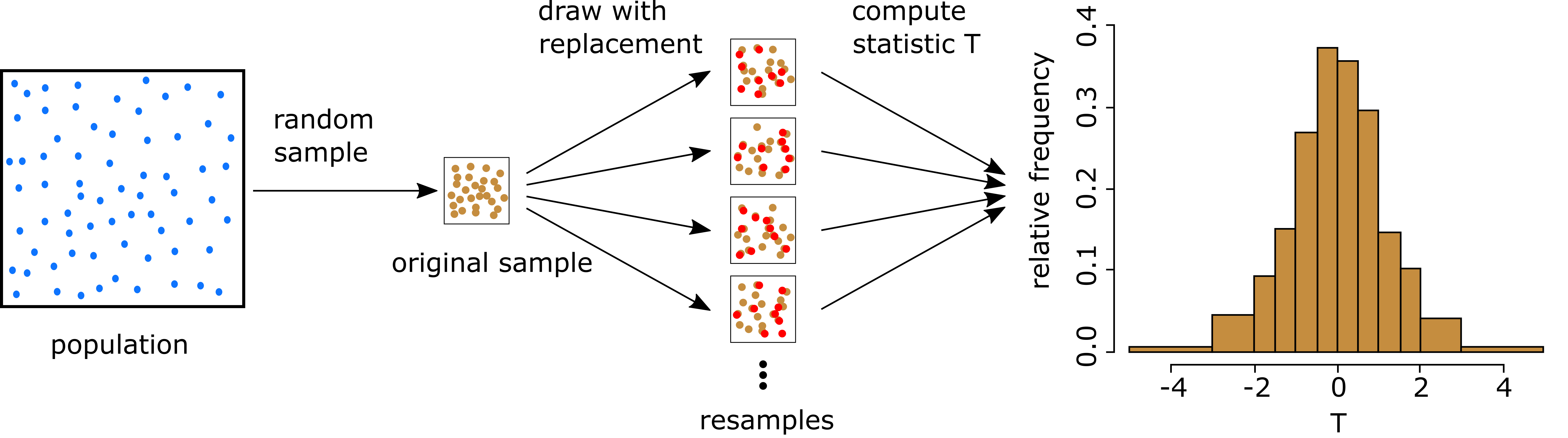
Setting: Assume given the original sample x_1,\ldots,x_n from unknown population f
Bootstrap: Regard the sample as the whole population
Replace the unknown distribution f with the sample distribution \hat{f}(x) = \begin{cases} \frac1n & \quad \text{ if } \, x \in \{x_1, \ldots, x_n\} \\ 0 & \quad \text{ otherwise } \end{cases}
Any sampling will be done from \hat{f} \qquad \quad (motivated by Glivenko-Cantelli Thm)
Note: \hat{f} puts mass \frac1n at each sample point
Drawing an observation from \hat f is equivalent to drawing one point at random from the original sample \{x_1,\ldots,x_n\}